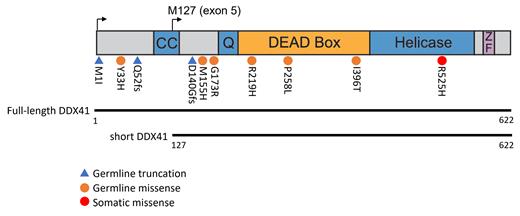
Germline variants in DDX41 are the most common cause of inherited predisposition to Myelodysplastic Syndromes (MDS) and Acute Myeloid Leukemia (AML). DDX41 codes for an RNA helicase with many described functions, and thus the precise role of these variants in disease is uncertain. Approximately 2/3 of disease-associated DDX41 variants cause truncation of the DDX41 protein due to frameshift, loss of a translation start, or splice alterations, while the other 1/3 of DDX41 variants are missense. The effect of these missense variants on DDX41 function is largely unknown and thus they are typically categorized as variants of unknown significance (VUSs). Importantly, 30-50% of MDS/AML patients with either truncating or missense DDX41 variants acquire a somatic mutation in the other allele of DDX41, and this mutation has strong preference for the amino acid substitution R525H. Our previous work demonstrated that the R525H mutation causes loss of DDX41 function. To better understand how germline variants affect DDX41 function, we designed a genetic complementation strategy in immortalized hematopoietic progenitor cells with inducible deletion of DDX41. We screened six of the most common missense VUSs by expressing them from a lentivirus in DDX41-proficient cells and then inducing deletion of endogenous DDX41 to determine the effect of each VUS on proliferation and survival of the cells. Empty vector-transduced DDX41-deleted cells undergo cell-cycle arrest and apoptosis within 3-4 days. In contrast, viral expression of wild-type DDX41 rescues the survival and proliferation of the cells, while expression of the R525H mutant fails to rescue the cells. We screened the Y33H, M155H, G173R, R219H, P258L, and I396T variants of DDX41 in this assay, and all six of the mutants successfully rescued the survival and proliferation of DDX41-deleted cells long-term. This indicates that the mutant proteins coded for by these variants are functional. To test the effect of commonly observed truncating variants in the same assay, we expressed M1I, Q52fs, and D140Gfs variants of DDX41. Surprisingly, we saw complete rescue of cell proliferation and survival by the M1I and Q52fs variants. In contrast, the D140Gfs variant failed to rescue the cells. These data indicate that frameshift at D140 or later causes loss of DDX41 function. It is likely that translation from alternative methionine residues located at M127 or M132 of DDX41 accounts for the rescue of the cells by truncating variants affecting sites upstream of M127. In support of this theory, we targeted DDX41 for CRISPR-mediated knockout in MOLM13 cells and found that guide RNAs targeting exons 1-4, which are upstream of M127, produced homozygous frameshift mutations in proliferating cells from single-cell clones. However, guides targeting exon 6, which is downstream of M127 and M132, did not produce any surviving cells with homozygous frameshift mutations, despite screening 96 clones. This finding indicates that short DDX41 isoforms expressed from alternative start sites are functional and can support cell proliferation when full-length protein is lost. Collectively, these data raise important questions about the precise role of DDX41 variants in hematologic malignancy. Data from recently published patient cohorts indicates that patients with missense DDX41 variants have similar patient characteristics to those with truncating variants and thus these aberrations are thought to cause disease by similar mechanisms. While these cell line models may not fully recapitulate the pathogenic mechanism by which DDX41 variants cause MDS/AML predisposition, the results of this study demonstrate that the germline variants are not complete loss-of-function alleles in all cases and more complex mechanisms may be involved.
Disclosures
Patnaik:Epigenetix: Research Funding; Kura: Research Funding; CTI BioPharma: Membership on an entity's Board of Directors or advisory committees; StemLine: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal